There’s been a lot of water under the bridge since the first release of Nextflow in July 2013. From its humble beginnings at the Centre for Genomic Regulation (CRG) in Barcelona, Nextflow has evolved from an upstart workflow orchestrator to one of the most consequential projects in open science software (OSS). Today, Nextflow is downloaded 120,000+ times monthly, boasts vibrant user and developer communities, and is used by leading pharmaceutical, healthcare, and biotech research firms.
On the occasion of Nextflow’s anniversary, I thought it would be fun to share some perspectives and point out how far we’ve come as a community. I also wanted to recognize the efforts of Paolo Di Tommaso and the many people who have contributed enormous time and effort to make Nextflow what it is today.
Bill Gates is credited with observing that “people often overestimate what they can do in one year, but underestimate what they can do in ten.” The lesson, of course, is that real, meaningful change takes time. Progress is measured in a series of steps. Considered in isolation, each new feature added to Nextflow seems small, but they combine to deliver powerful capabilities.
Life sciences has seen a staggering amount of innovation. According to estimates from the National Human Genome Research Institute (NHGRI), the cost of sequencing a human genome in 2013 was roughly USD 10,000. Today, sequencing costs are in the range of USD 200—a 50-fold reduction.1
A fundamental principle of economics is that “if you make something cheaper, you get more of it.” One didn’t need a crystal ball to see that, driven by plummeting sequencing and computing costs, the need for downstream analysis was poised to explode. With advances in sequencing technology outpacing Moore’s Law, It was clear that scaling analysis capacity would be a significant issue.2
When Paolo and his colleagues started the Nextflow project, it was clear that emerging technologies such as cloud computing, containers, and collaborative software development would be important. Even so, it is still amazing how rapidly these key technologies have advanced in ten short years.
In an article for eLife magazine in 2021, Paolo described how Solomon Hyke’s talk “Why we built Docker” at DotScale in the summer of 2013 impacted his thinking about the design of Nextflow. It was evident that containers would be a game changer for scientific workflows. Encapsulating application logic in self-contained, portable containers solved a multitude of complexity and dependency management challenges — problems experienced daily at the CRG and by many bioinformaticians to this day. Nextflow was developed concurrent with the container revolution, and Nextflow’s authors had the foresight to make containers first-class citizens.
With containers, HPC environments have been transformed — from complex environments where application binaries were typically served to compute nodes via NFS to simpler architectures where task-specific containers are pulled from registries on demand. Today, most bioinformatic pipelines use containers. Nextflow supports multiple container formats and runtimes, including Docker, Singularity, Podman, Charliecloud, Sarus, and Shifter.
Some of the earliest efforts around Nextflow centered on building high-quality executors for HPC workload managers. A key idea behind schedulers such as LSF, PBS, Slurm, and Grid Engine was to share a fixed pool of on-premises resources among multiple users, maximizing throughput, efficiency, and resource utilization.
See the article Nextflow on BIG IRON: Twelve tips for improving the effectiveness of pipelines on HPC clusters
While cloud infrastructure was initially “clunky” and hard to deploy and use, the idea of instant access and pay-per-use models was too compelling to ignore. In the early days, many organizations attempted to replicate on-premises HPC clusters in the cloud, deploying the same software stacks and management tools used locally to cloud-based VMs.
With the launch of AWS Batch in December 2016, Nextflow’s developers realized there was a better way. In cloud environments, resources are (in theory) infinite and just an API call away. The traditional scheduling paradigm of sharing a finite resource pool didn’t make sense in the cloud, where users could dynamically provision a private, scalable resource pool for only the duration of their workload. All the complex scheduling and control policies that tended to make HPC workload managers hard to use and manage were no longer required.3
AWS Batch also relied on containerization, so it only made sense that AWS Batch was the first cloud-native integration to the Nextflow platform early in 2017, along with native support for S3 storage buckets. Nextflow has since been enhanced to support other batch services, including Azure Batch and Google Cloud Batch, along with a rich set of managed cloud storage solutions. Nextflow’s authors have also embraced Kubernetes, developed by Google, yet another way to marshal and manage containerized application environments across public and private clouds.
A major trend shaping software development has been the use of collaborative source code managers (SCMs) based on Git. When Paolo was thinking about the design of Nextflow, GitHub had already been around for several years, and DevOps techniques were revolutionizing software. These advances turned out to be highly relevant to managing pipelines. Ten years ago, most bioinformaticians stored copies of pipeline scripts locally. Nextflow’s authors recognized what now seems obvious — it would be easier to make Nextflow SCM aware and launch pipelines directly from a code repository. Today, this simple idea has become standard practice. Most users run pipelines directly from GitHub, GitLab, Gitea, or other favorite SCMs.
A few basic concepts and patterns in computer science appear repeatedly in different contexts. These include iteration, indirection, abstraction, and component reuse/modularization. Enabled by containers, we have seen a significant shift towards modularization in bioinformatics pipelines enabled by catalogs of reusable containers. In addition to general-purpose registries such as Docker Hub and Quay.io, domain-specific efforts such as biocontainers have emerged, aimed at curating purpose-built containers to meet the specialized needs of bioinformaticians.
We have also seen the emergence of platform and language-independent package managers such as Conda. Today, almost 10,000 Conda recipes for various bioinformatics tools are freely available from Bioconda. Gone are the days of manually installing software. In addition to pulling pre-built bioinformatics containers from registries, developers can leverage packages of bioconda recipes directly from the bioconda channel.
The Nextflow community has helped lead this trend toward modularization in several areas. For example, in 2022, Seqera Labs introduced Wave. This new service can dynamically build and serve containers on the fly based on bioconda recipes, enabling the two technologies to work together seamlessly and avoiding building and maintaining containers by hand.
With nf-core, the Nextflow community has extended the concept of modularization and reuse one step further. Much as bioconda and containers have made bioinformatics software modular and portable, nf-core modules extend these concepts to pipelines. Today, there are 900+ nf-core modules — essentially building blocks with pre-defined inputs and outputs based on Nextflow’s elegant dataflow model. Rather than creating pipelines from scratch, developers can now wire together these pre-assembled modules to deliver new functionality rapidly or use any of 80 of the pre-built nf-core analysis pipelines. The result is a dramatic reduction in development and maintenance costs.
Since the first Nextflow release in July 2013, there have been 237 releases and 5,800 commits. Also, the project has been forked over 530 times. There have been too many important enhancements and milestones to capture here. We capture some important developments in the timeline below:
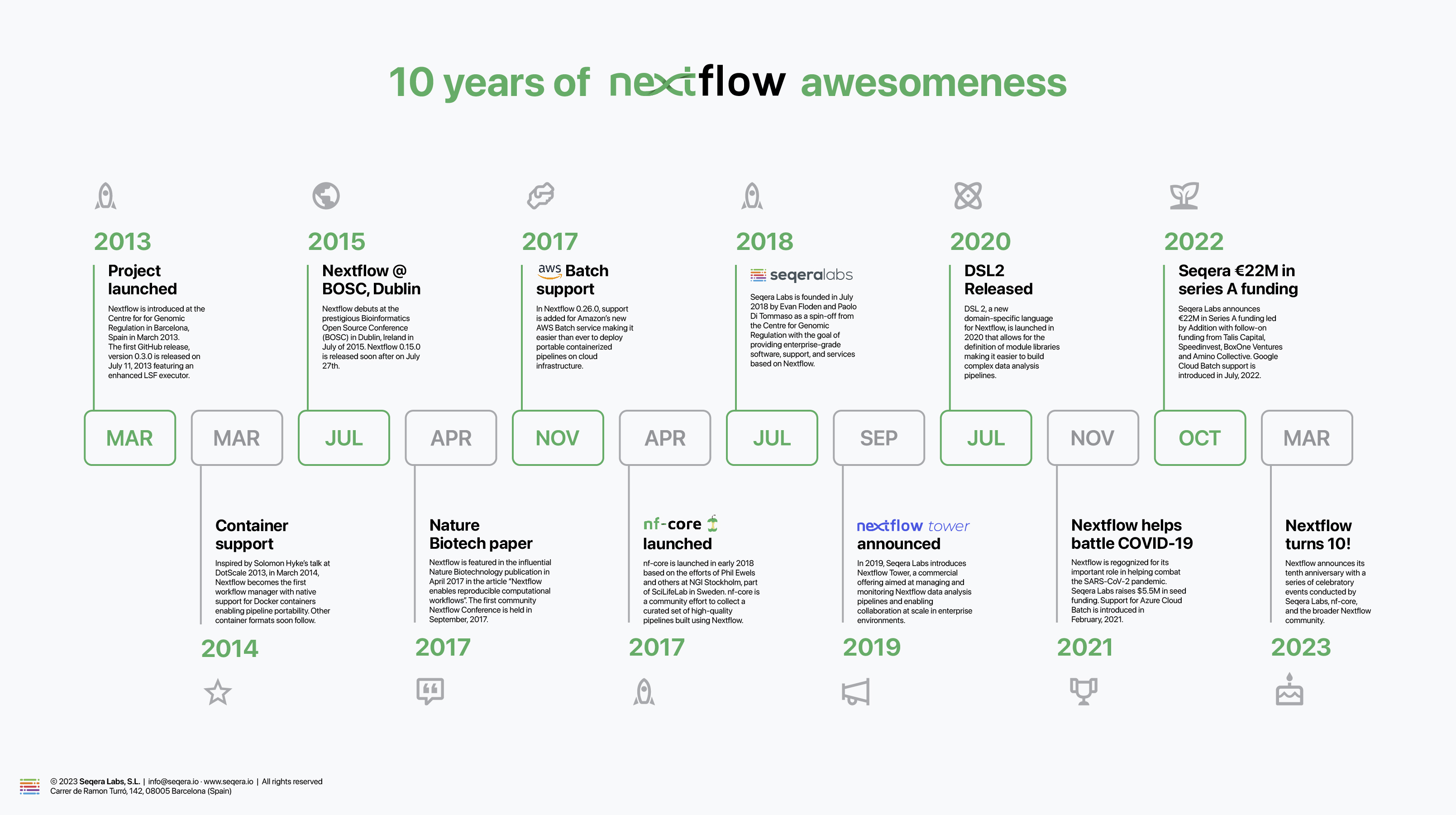
As we look to the future, the pace of innovation continues to increase. It’s been exciting to see Nextflow expand beyond the various omics disciplines to new areas such as medical imaging, data science, and machine learning. We continue to evolve Nextflow, adding new features and capabilities to support these emerging use cases and support new compute and storage environments. I can hardly wait to see what the next ten years will bring.
For those new to Nextflow and wishing to learn more about the project, we have compiled an excellent collection of resources to help you Learn Nextflow in 2023.
1 https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost 2 Coined by Gordon Moore of Intel in 1965, Moore’s Law predicted that transistor density, roughly equating to compute performance, would roughly double every two years. This was later revised in some estimates to 18 months. Over ten years, Moore’s law predicts roughly a 2^5 = 32X increase in performance – less than the ~50-fold decrease in sequencing costs. See chart here. 3 This included features like separate queues, pre-emption policies, application profiles, and weighted fairshare algorithms.